Difference between revisions of "Virtual Screening"
(→Virtual Screening Manual) |
(→Job monitoring) |
||
| Line 38: | Line 38: | ||
== Job monitoring == | == Job monitoring == | ||
<p>On Step 6 screen, users have the option to type in their jobid, check status and get results.</p> | <p>On Step 6 screen, users have the option to type in their jobid, check status and get results.</p> | ||
| − | [[Image: | + | [[Image:vs7.png|900px|center]] |
<p><center><strong>Figure 6. </strong> job monitoring screenshot | <p><center><strong>Figure 6. </strong> job monitoring screenshot | ||
</center></p> | </center></p> | ||
Revision as of 08:01, 8 November 2021
Virtual Screening Manual
VSTH can provide a complete workflow for molecular docking, including PDB file preparation, pocket setting, molecular database preparation, docking program selection, job monitoring, and result analysis and visualization.
Target protein preparation
For processing protein, the end users can follow the prompts. The protein target can be accessed by either providing a 4-character PDB accession code (1) or by uploading a PDB file (2). Users also can remove waters, ions, ligands in PDB files (3) as they want. Besides, users can hydrogenate proteins (4). There are 3 methods for adding hydrogen implemented in REDUCE package:
- First, add H rotate and flip NQH groups.
- Second, add H and rotate groups with no NQH flips.
- Third, add H, including His sc NH, then rotate and flip groups.

Pocket setting
VSTH can automatically identify pockets using Fpocket package and present an interactive viewer of the docking site with the highest score(1). Detail information on the provided target (2), such as druggability score, number of alpha spheres, total SASA and so on, can also be found on this package. Users can select the top 10 identified pockets by Fpocket package or manually change the box size and center for the pocket (3). What’s more, users can define the number of poses (4) for the selected docking programs.

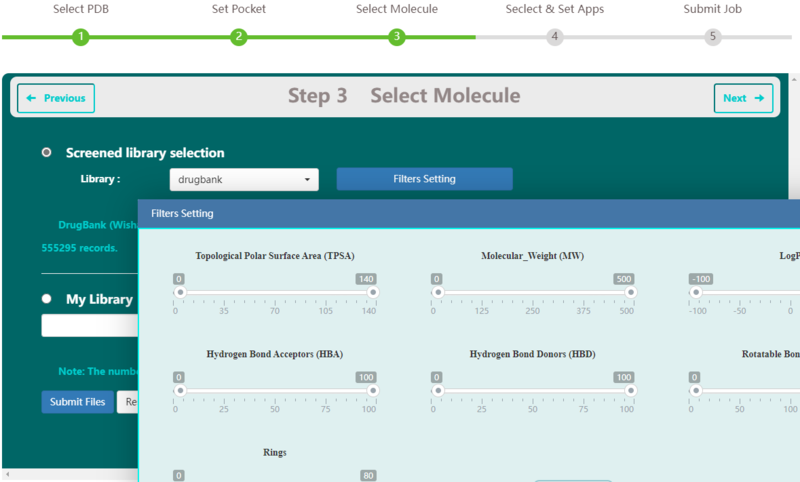
Ligand library preparation
Step 3 allows the user to select libraries for virtual screening on VSTH. VSTH provides one public ligand library and one test ligand library. The data of the public library is selected randomly from ZINC15 (Stering, et al, 2015) and 1000 records are included. Users can also upload their own ligand library. VSTH supports compressed files, such as zip file, tar file, and so on. Also, VSTH uses OpenBabel (O’Boyle et al., 2011) for format transformation, so VSTH supports formats like smi, mol2, mol, xyz, cif and sdf.
Docking program selection
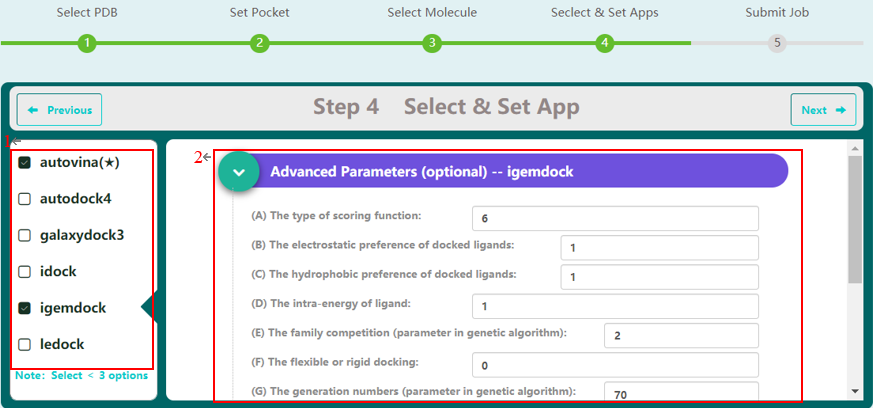
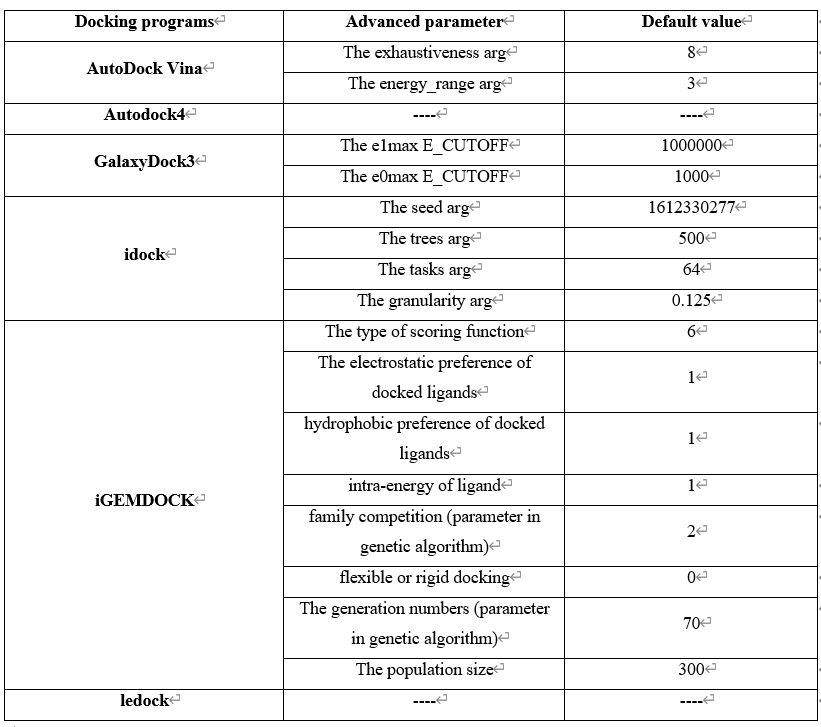
Step 4 allows the user to select docking programs and setting parameters for virtual screening on VSTH. VSTH provides 6 software for docking (1). Users can select more than 1 program for docking. Generally, VSTH provides default parameters for docking, but users can modify parameters related to docking (2), such as the type of scoring function, the electrostatic preference of docked ligands, on this screen. All advanced parameters are provided according to docking programs and can be found in Table 2. Besides these advanced parameters, users have the option to select DLIGAND2 to predict protein-ligand binding affinity.
Job submission
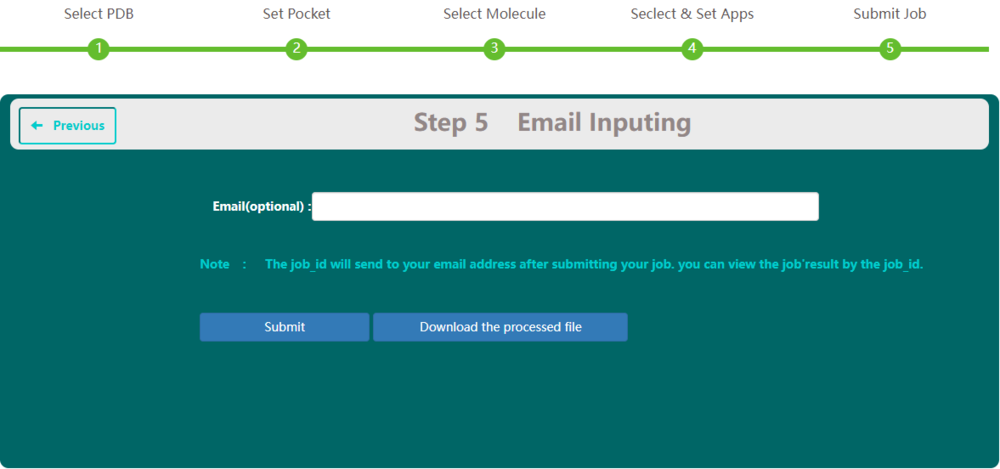
On Step 5 screen, users have the option to type in their email. This email address will be used to receive taskid. This taskid is a unique identifier, and users need it in the job monitoring process to check its status and get results.
Job monitoring
On Step 6 screen, users have the option to type in their jobid, check status and get results.