Virtual Screening
Virtual Screening Manual
VSTH can provide a complete workflow for molecular docking, including PDB file preparation, pocket setting, molecular database preparation, docking program selection, job monitoring, and result analysis and visualization.By using this website, you agree that VSTH will not be liable for any losses or damages arising from your use of or reliance on the Content, or other websites or information to which this website may be linked.
Target protein preparation
During the input file preparation process, the end user can follow the prompts on the web page to provide a structure of the protein target by either providing a PDB code (1) or by uploading a PDB file (2). By clicking the eye icon, users can show or hide protein, ligand, water, ion. By clicking the delete icon, users also can delete water, ion, ligand and protein. By clicking the submenu icon, users can find the detail list of selected category. For example, if users clicked the submenu icon of protein, it will show its chain list. What’s more, users can add hydrogen to the target protein (4), there are 3 types of adding hydrogen:
- add H rotate and flip NQH groups.
- add H and rotate groups with no NQH flips.
- add H, including His sc NH, then rotate and flip groups.
Pocket setting
VSTH will automatically identify pockets using fpocket [9] and present an interactive viewer of the protein target and the best scoring pocket (1). Detailed information on the provided target (3), such as drug ability score, number of alpha spheres, total SASA and so on, can also be found on this page. Users can select the top 10 identified pockets by fpocket or input the size and center of pocket directly (2). What’s more, users can define the number of poses (4)(Fig. 2).

Ligand library preparation
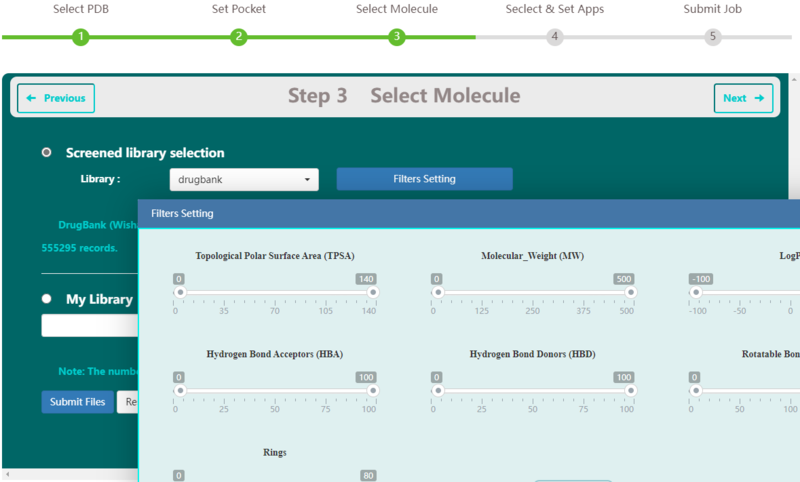
Step 3 allows the user to select libraries. Available public libraries in VSTH include DrugBank (Wishart D. S., 2006), HMDB (Wishart D. S., 2007), and IBS (Ltd., 2021). DrugBank contains 2387 records. HMDB contains 113875 records. And IBS contains 555295 records. Users can set filters to these libraries or upload their own ligand libraries. VSTH supports compressed files and other file formats like smi, mol2, mol, xyz, cif and sdf, etc. (Fig. 3).
Docking program selection
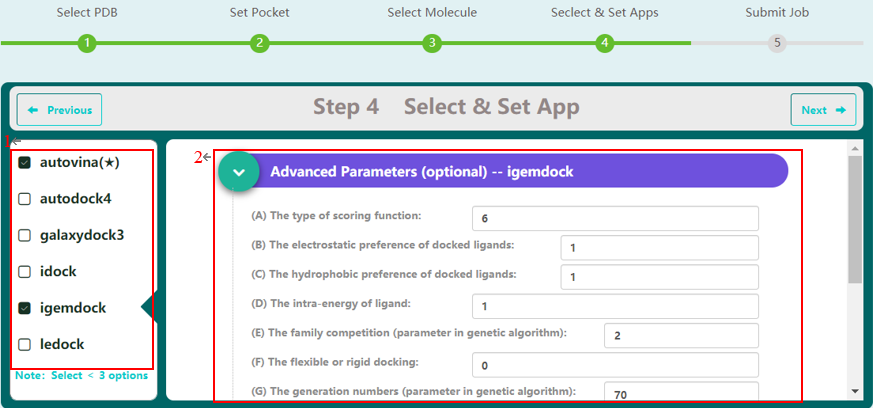
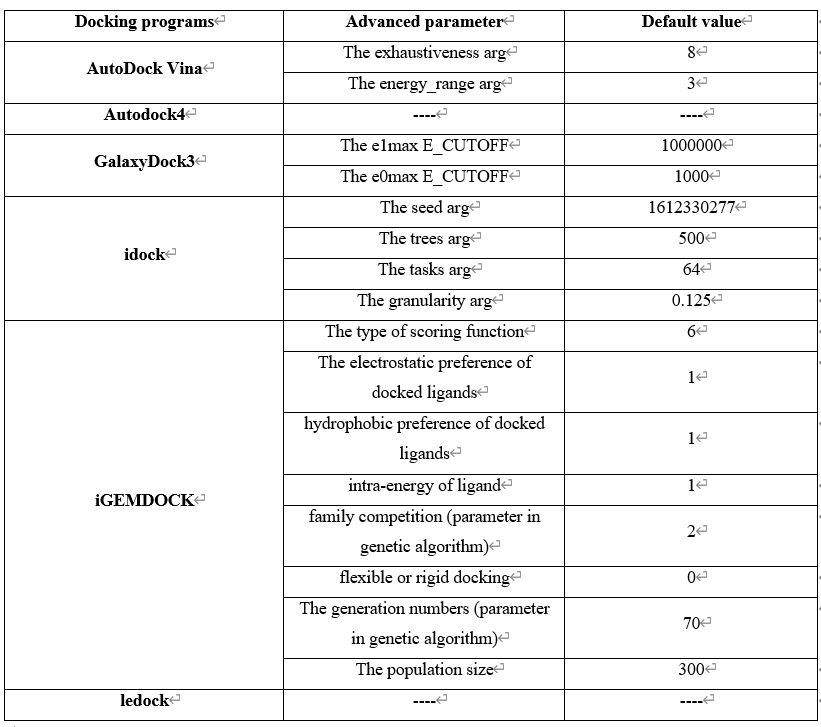
Step 4 allows the user to select docking programs and set parameters. VSTH provides 6 docking programs (1). Users can select at most 3 programs at one time. Generally, VSTH provides default parameters for docking, while users can modify parameters for personalized docking (2). All advanced parameters are provided according to docking programs and can be found in Table 2. Besides these advanced parameters, users have the option to select DLIGAND2 to re-score the docking conformations. For conformation classification, users can set the cutoff argument (Fig. 4).
Job submission
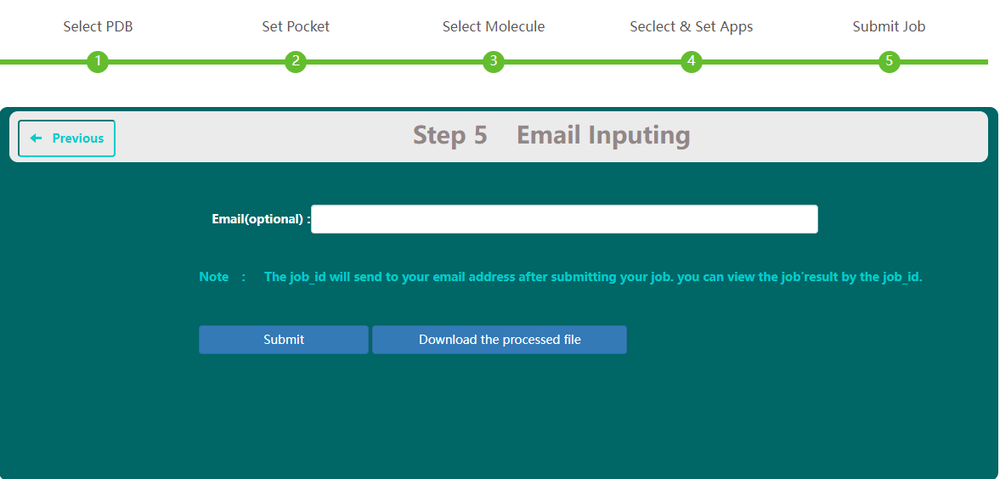
On Step 5, users have the option to type in their email. This email address will be used to receive the task id and get reminder when the task has been finished or cancelled.And it will be erased as soon as you received your results
Job monitoring
On Step 6 screen, users have the option to type in their task id, check status, get results and cancel task.
FAQ
1. email
==2. environment==